Transcriptional regulation of tissue-resident memory cells
Maximilian Heeg
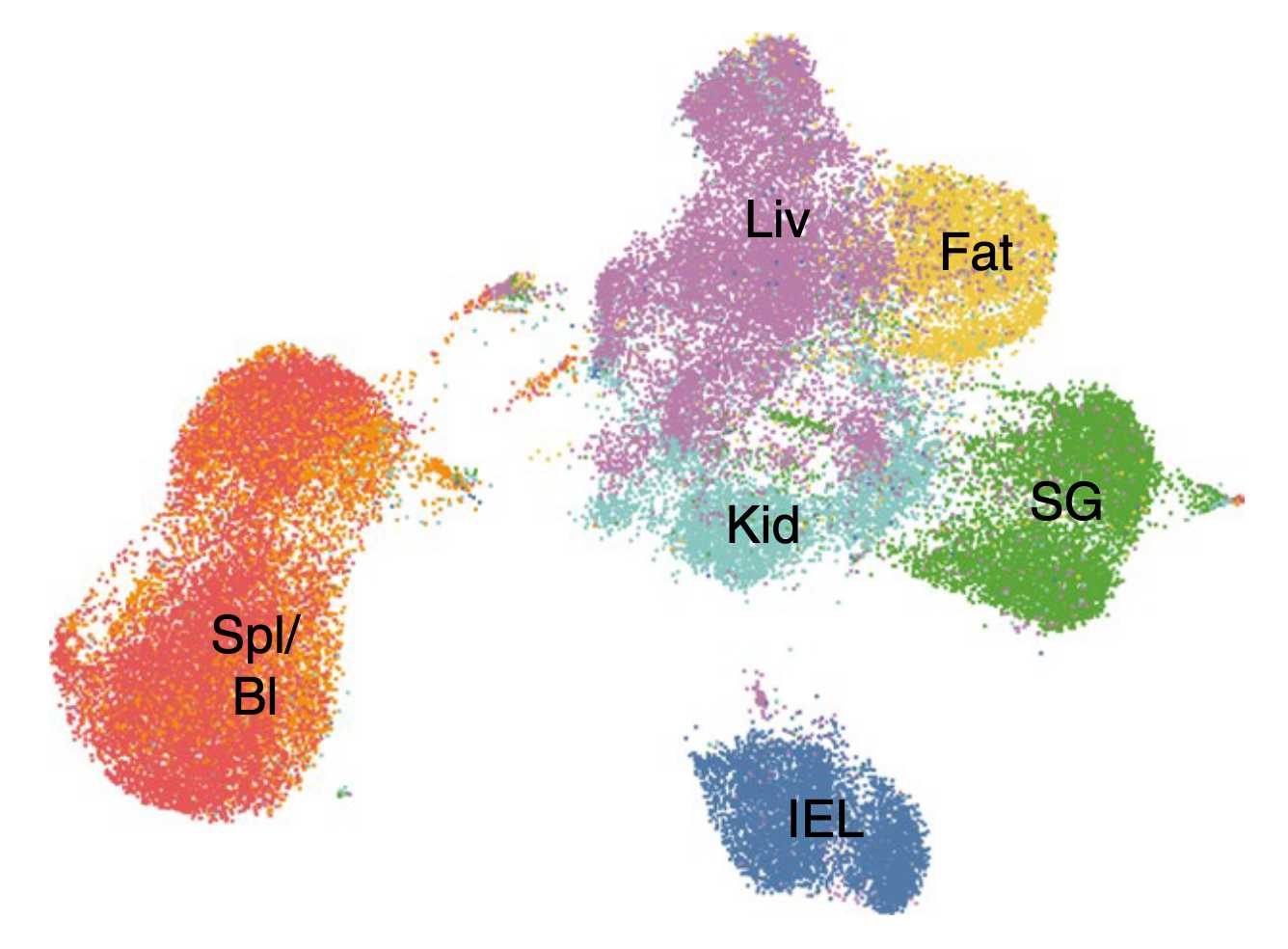
How do tissue-resident memory cells adapt to unique tissue microenvironments? How do they sense environmental signals? How are they incorporated? Using mouse models of acute viral infection, combined with genetic perturbations and single-cell sequencing, we explore the transcriptional networks that govern the acclimatization of T cells to various barrier tissues.
Tissue-resident memory CD8+ T cells (TRM) localize to infected barrier tissues like the intestine and provide a local first line of defense against reinfection. However, TRM cells are also associated with uncontrolled immune activation, manifesting in diseases like inflammatory bowel disease, or graft-versus-host-disease. As TRM reside in many tissues that may differ in microenvironment, including the cytokine milieu and cellular composition, they require tissue-specific acclimatization mechanisms to persist in and patrol these tissues.
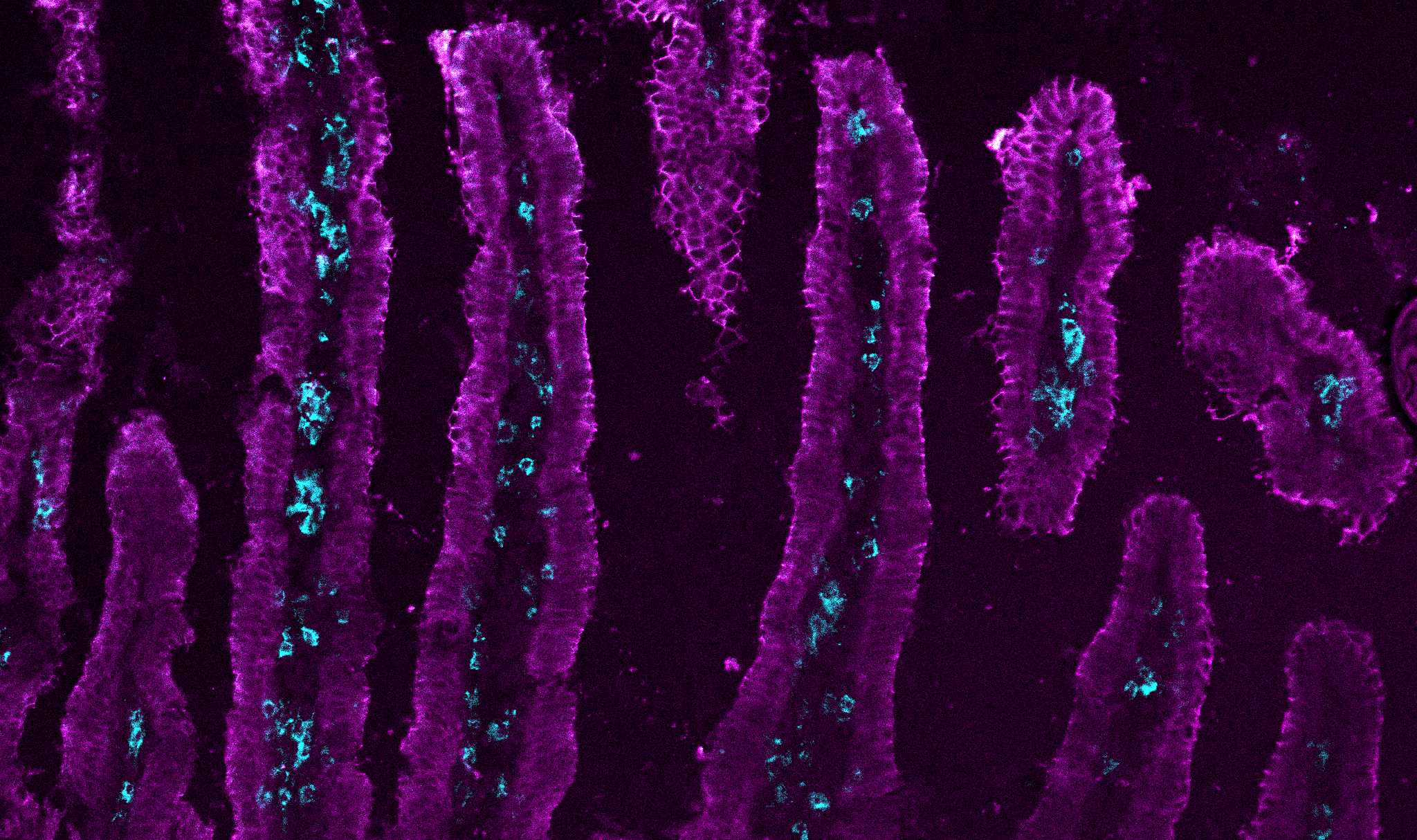
Immunofluorescence of T cells in the small intestine after LCMV infection.
A complex network of transcription factors regulates the differentiation of normadic circulating T cells into resident memory cells. We have recently identified the transcription factor Hic1 as a specific regulator for small intestinal T cells. The questions that we are currently trying to answer are: Which cues and cellular interactions regulate these adaptations? How are transcriptional circuits controlling tissue residency, immune function, and memory potential integrated? What are the specific targets of Hic1 and its mechanisms of action, and how are these adaptations regulated in sustained infection or tumors?
Related Publications
- Nat Immunol. 20222022/tissue-resident-memory-cd8-t-cells-possess-unique-transcriptional-epigenetic-and-functional-adaptations-to-different-tissue-environments