Spatial Orchestration of small intestinal tissue-resident T cells
Maximilian Heeg
Using spatial transcriptomics, we try to overcome limiations of single-cell sequencing and study T cells in intact tissues to understand which cell-cell interactions, gradients and cellular niches promote memory formation in barrier tissues.
Many tools to study tissue-resident T cells necessarily rely on harsh isolation methods to separate the cells-of-interest from the surrounding tissue, thus depriving them of their environment. However, it has become increasingly clear that this isolation limits our understanding of how immune responses are regulated in tissues. During my postdoctoral work, I was able to show that spatial transcriptomics, which overcomes the limitation of cell isolation, can be used to study TRM cells in intact tissues. Indeed, preliminary data show, that much of the TRM cell heterogeneity that we observe within a single tissue, is, in fact, due to unique positioning of these TRM cells in relation to the tissue anatomy.
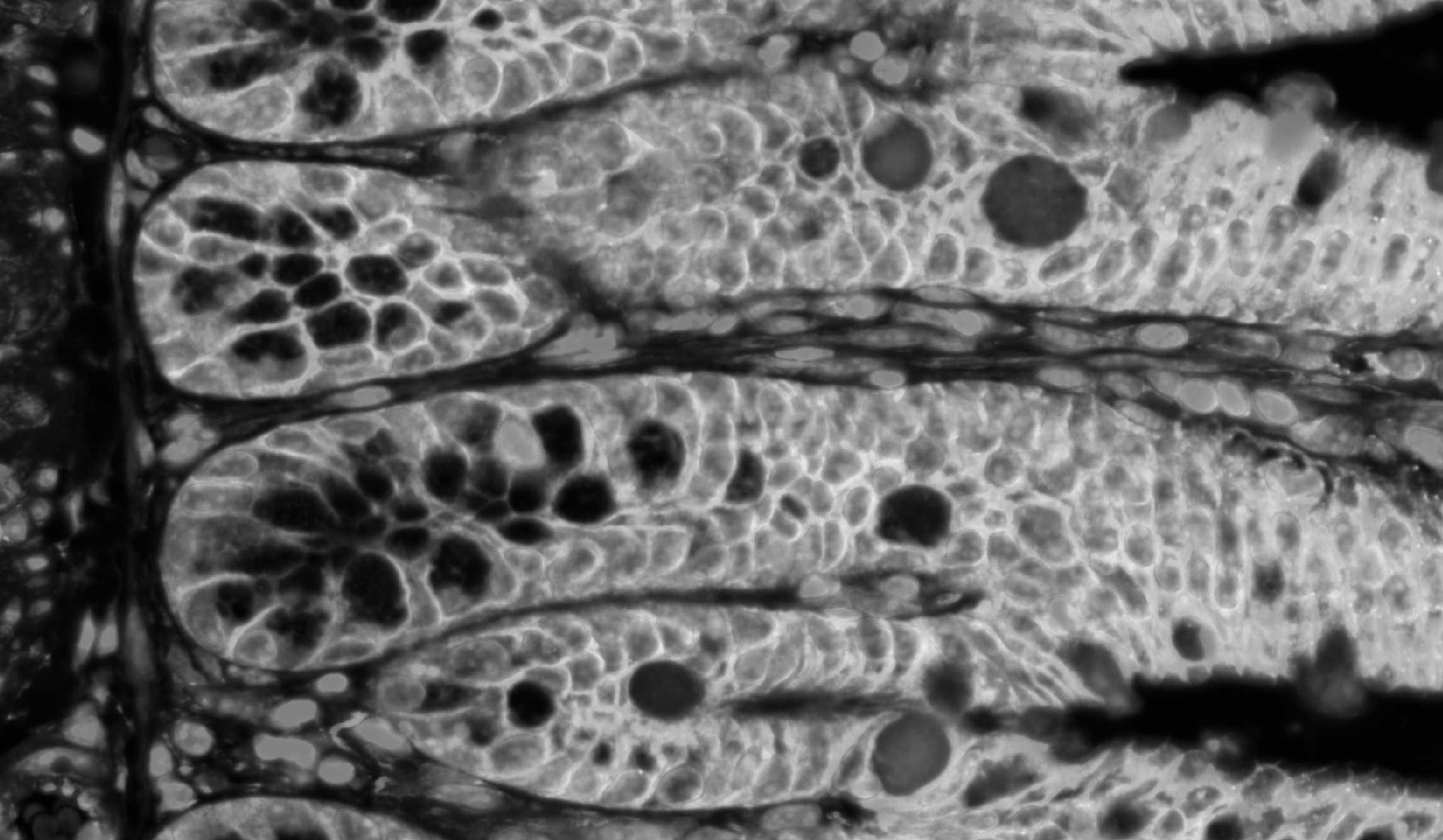
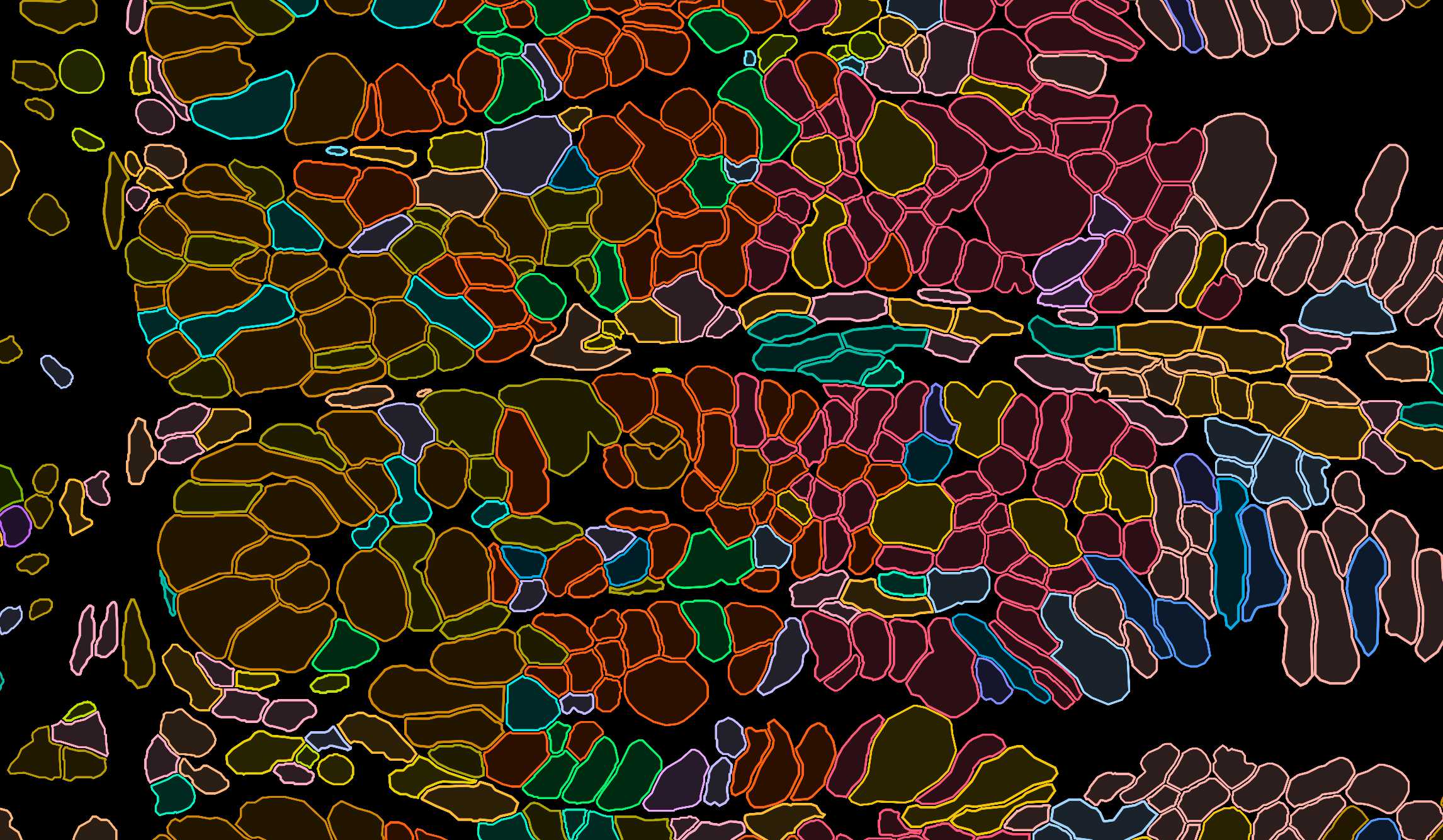
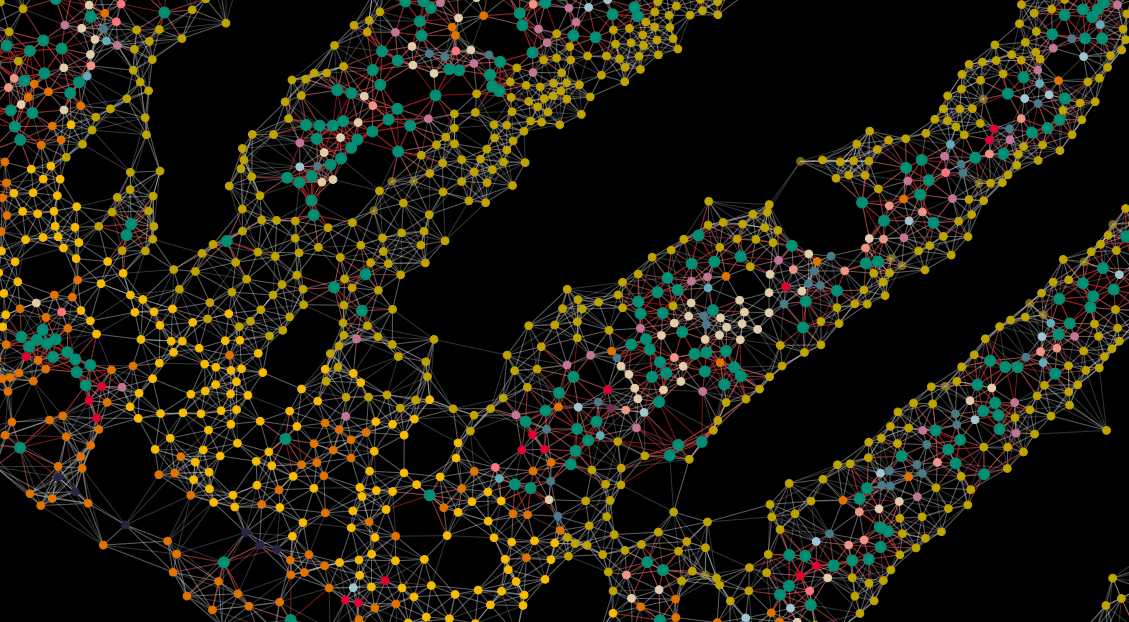
Xenium experiment of a mouse small intestine. Different layers of data quality are shown: Image data, cell and transcript data.
Spatial transcriptomics is a powerful, but challenging technique to study immune cells. While it allows to study T cells in undisrupted tissues, thereby allowing for the experiments outlined above, it comes with certain hurdles and limitations, that need to be overcome - ranging from designing a panel that captures the various cell types, but simultaneously gives sufficient resolution to annotate immune cell subtypes and states, improving and assessing cell segmentation and a cell annotation. Over the last years of my postdoc, I was able to develop a streamlined process to design targeted gene panels, perform cell segmentation, and impute missing genes from paired full-transcriptome single-cell RNA sequencing data. We have successfully used these tools, to study the diversity of TRM cells in the small intestine.
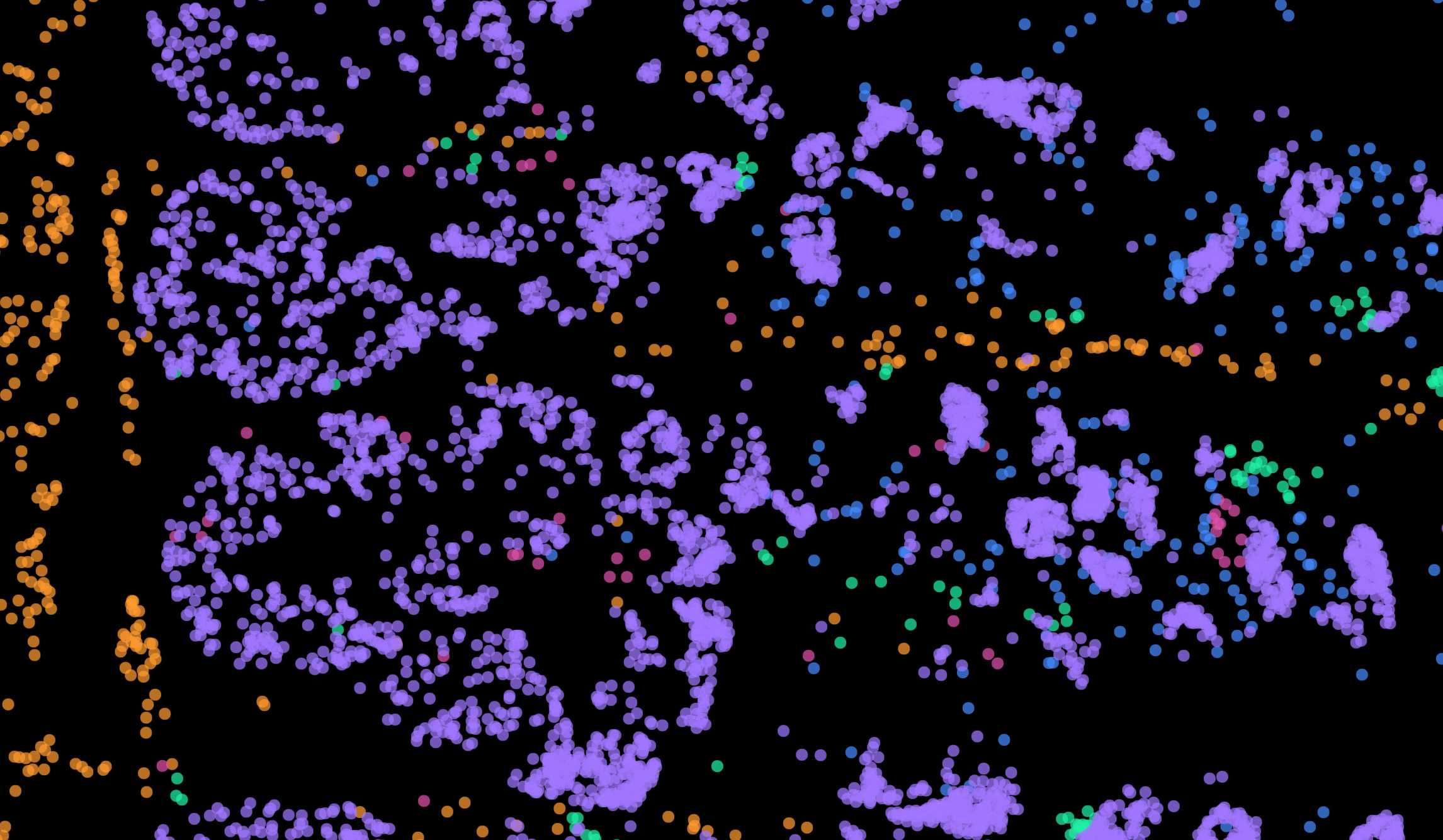
Spatial transcriptomics of the small intestine. Cells can be viewed in UMAP or spatial coordinates
Using this process, we found that TRM populations were spatially segregated: with more effector- and memory-like TRM preferentially localized at the villus tip or crypt, respectively. Modeling ligand-receptor activity revealed patterns of key cellular interactions and cytokine signaling pathways that initiate and maintain TRM differentiation and functional diversity, including different TGFβ sources. We proved that our framework can be used for the study of immune cell and revealed that T cell location and functional state are fundamentally intertwined
Related Publications
- Nature. 20252025/tissue-resident-memory-cd8-t-cell-diversity-is-spatiotemporally-imprinted