Projects
Current Research Focus
Our research aims to understand how immune responses are initiated, maintained, and regulated within tissues. By leveraging cutting-edge technologies and spatial transcriptomics, we investigate the signals and cell interactions that drive tissue adaptation, the transcription factors that regulate acclimatization to tissues, and how these processes can be therapeutically targeted.
Spatial Orchestration of small intestinal tissue-resident T cells
Using spatial transcriptomics, we try to overcome limiations of single-cell sequencing and study T cells in intact tissues to understand which cell-cell interactions, gradients and cellular niches promote memory formation in barrier tissues. [See how we use spatial transcriptomics to uncover the formation of TRM cells]
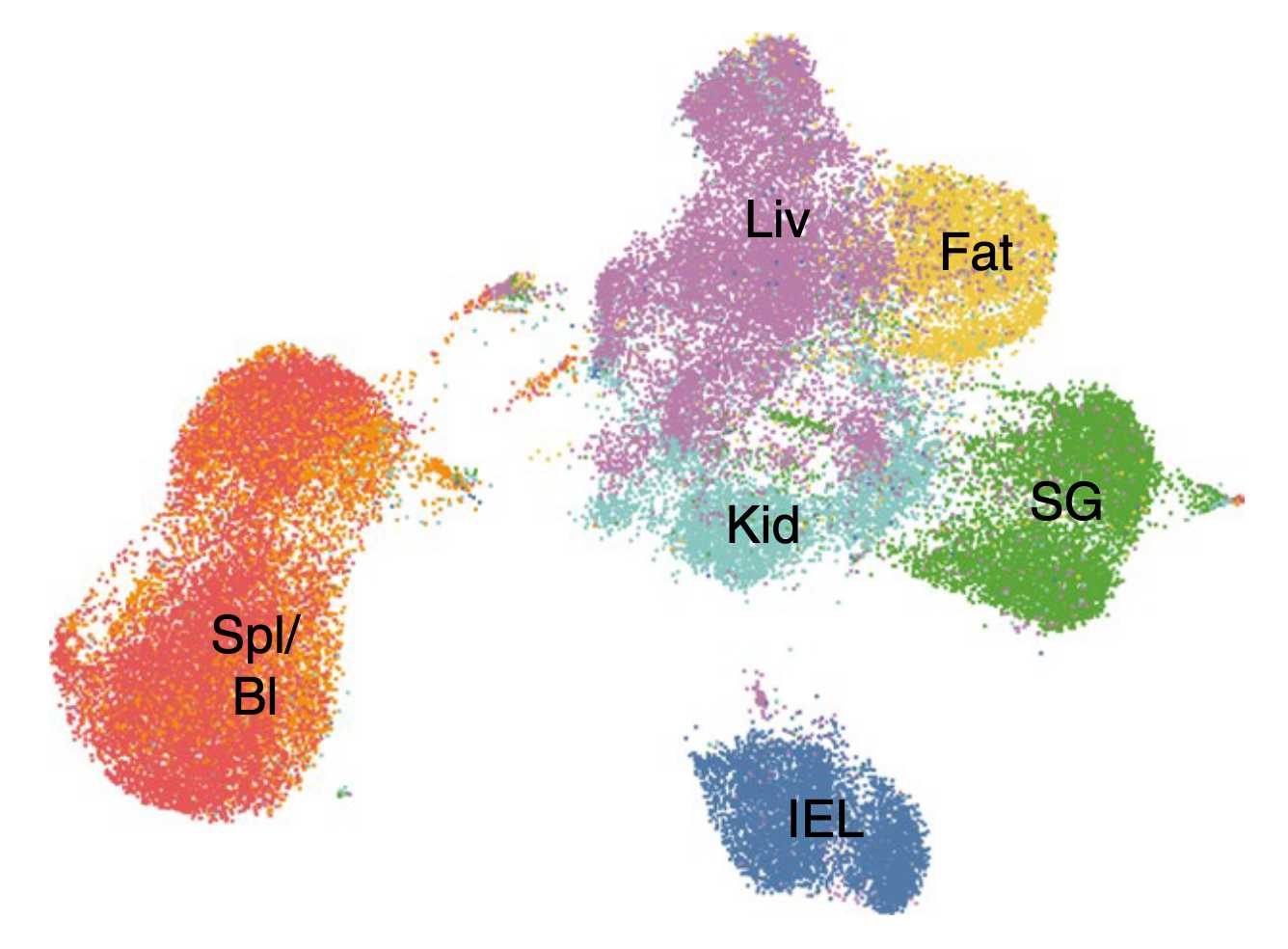
Transcriptional regulation of tissue-resident memory cells
How do tissue-resident memory cells adapt to unique tissue microenvironments? How do they sense environmental signals? How are they incorporated? Using mouse models of acute viral infection, combined with genetic perturbations and single-cell sequencing, we explore the transcriptional networks that govern the acclimatization of T cells to various barrier tissues. [Learn more about the transcriptional regulation of TRM cells]
Past Projects
Things that I worked on in the past and that sparked my interest in immunology.
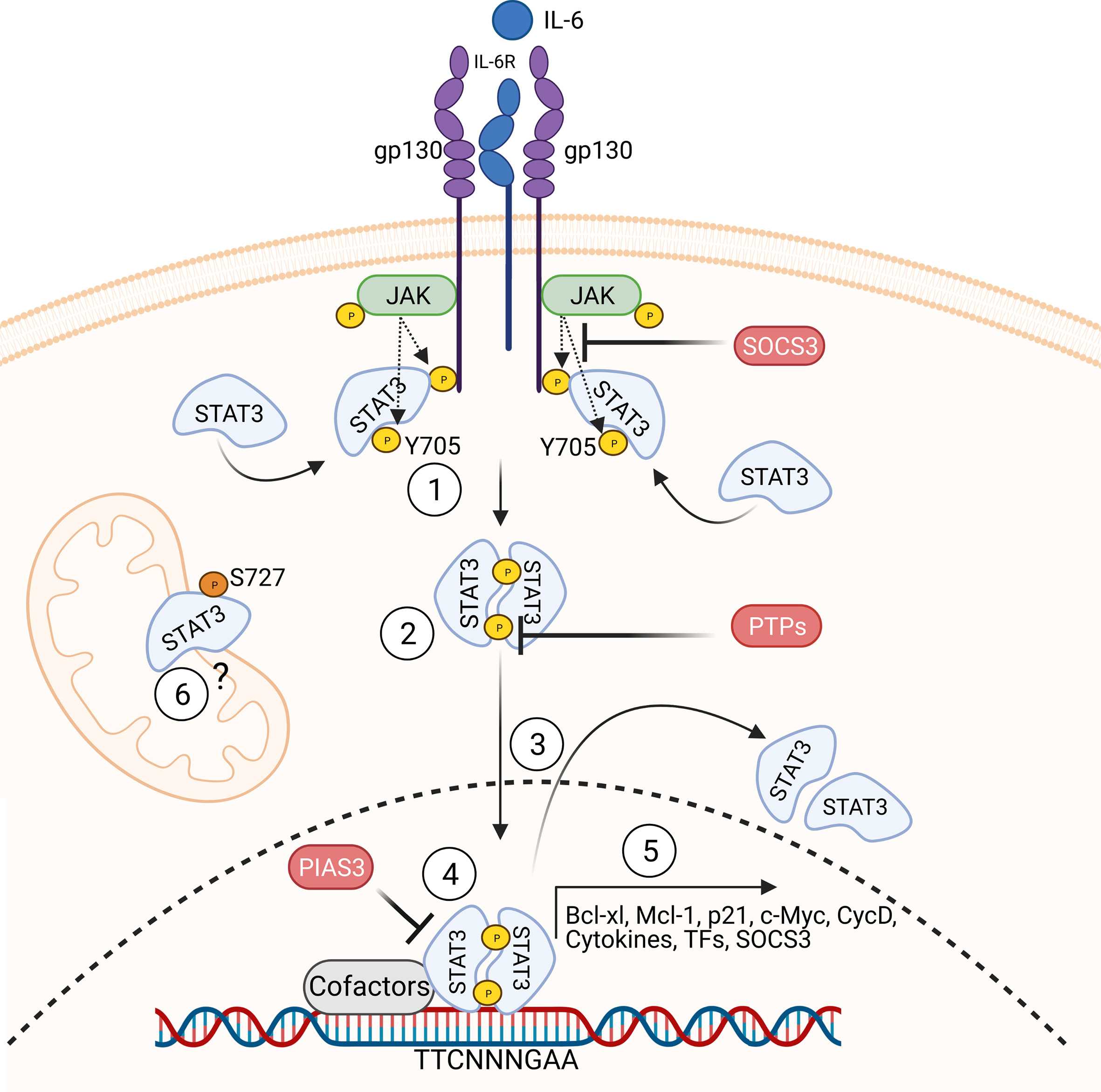
Molecular response patterns of germline STAT3 gain-of-function mutations
Primary immunodeficiencies provide a unique opportunity to study the human immune system, as germline mutations in patients can be directly and causally linked to their disease phenotype. Nevertheless, numerous inborn errors of immunity exhibit a significant degree of heterogeneity in the disease manifestation and penetrance. In this study, we categorized mutations in the STAT3 gene based on their molecular phenotype and associated them with the clinical penetrance and severity of the patients. [Read about the correlation of genotype and phenotype in STAT3 GOF]
Absence of infectious triggers in Hemophagocytic lymphohistiocytosis (HLH)
Hemophagocytic lymphohistiocytosis (HLH) is a severe hyperinflammatory condition that can arise from genetic mutations affecting the cytotoxic pathway, particularly in genes like PRF1, which are crucial for the function and regulation of cytotoxic T lymphocytes and NK cells. While HLH has traditionally been understood to be triggered by infections, most notably Epstein-Barr virus (EBV), our research across two distinct cohorts has revealed that HLH can manifest without an infectious trigger. The first cohort focused on CNS-isolated HLH cases, demonstrating that the disease can present in a localized manner rather than systemically. The second cohort examined newborns and cases of intrauterine HLH, where the disease developed before or immediately after birth, in the absence of infection exposure, providing compelling evidence that the genetic defect alone can be sufficient to initiate the inflammatory cascade characteristic of HLH. [Learn how HLH can be triggered without an infection]
The Importance of HBV-Specific CD8+ T Cells and Their Antiviral Efficacy
How do T cells contribute to viral control in HBV infection? Using a novel model system, we found that both cytolytic and non-cytolytic mechanisms were involved in the antiviral activity. [Learn more about how T cells fight HBV infections]